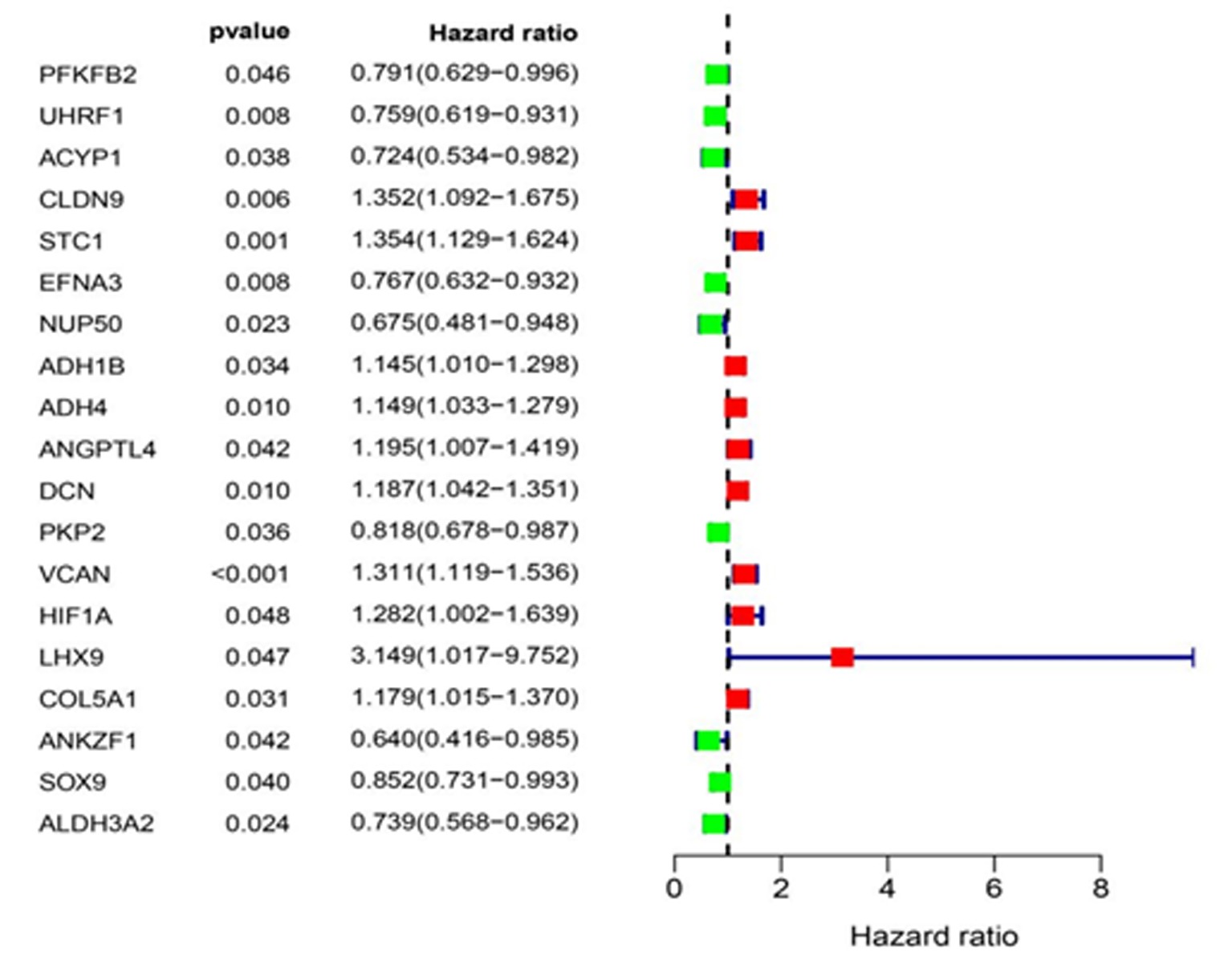
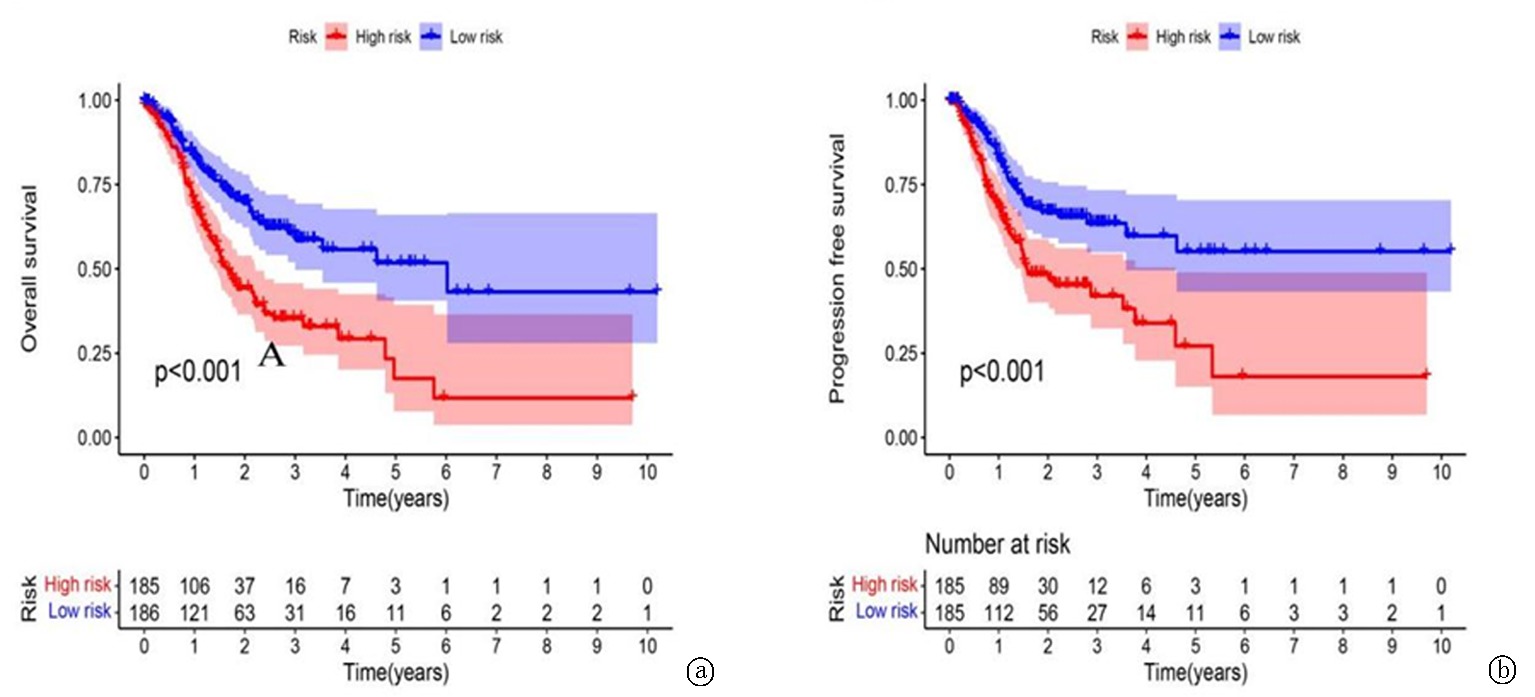
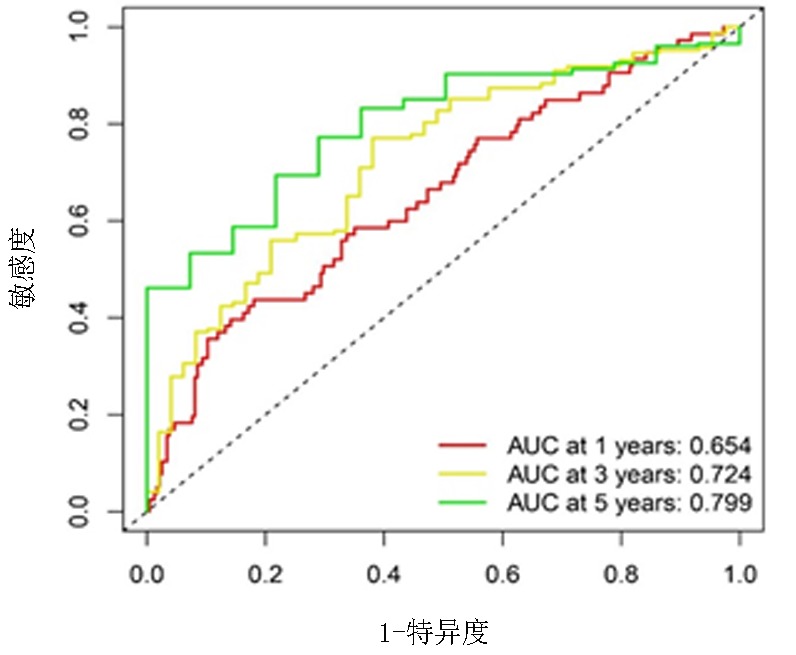
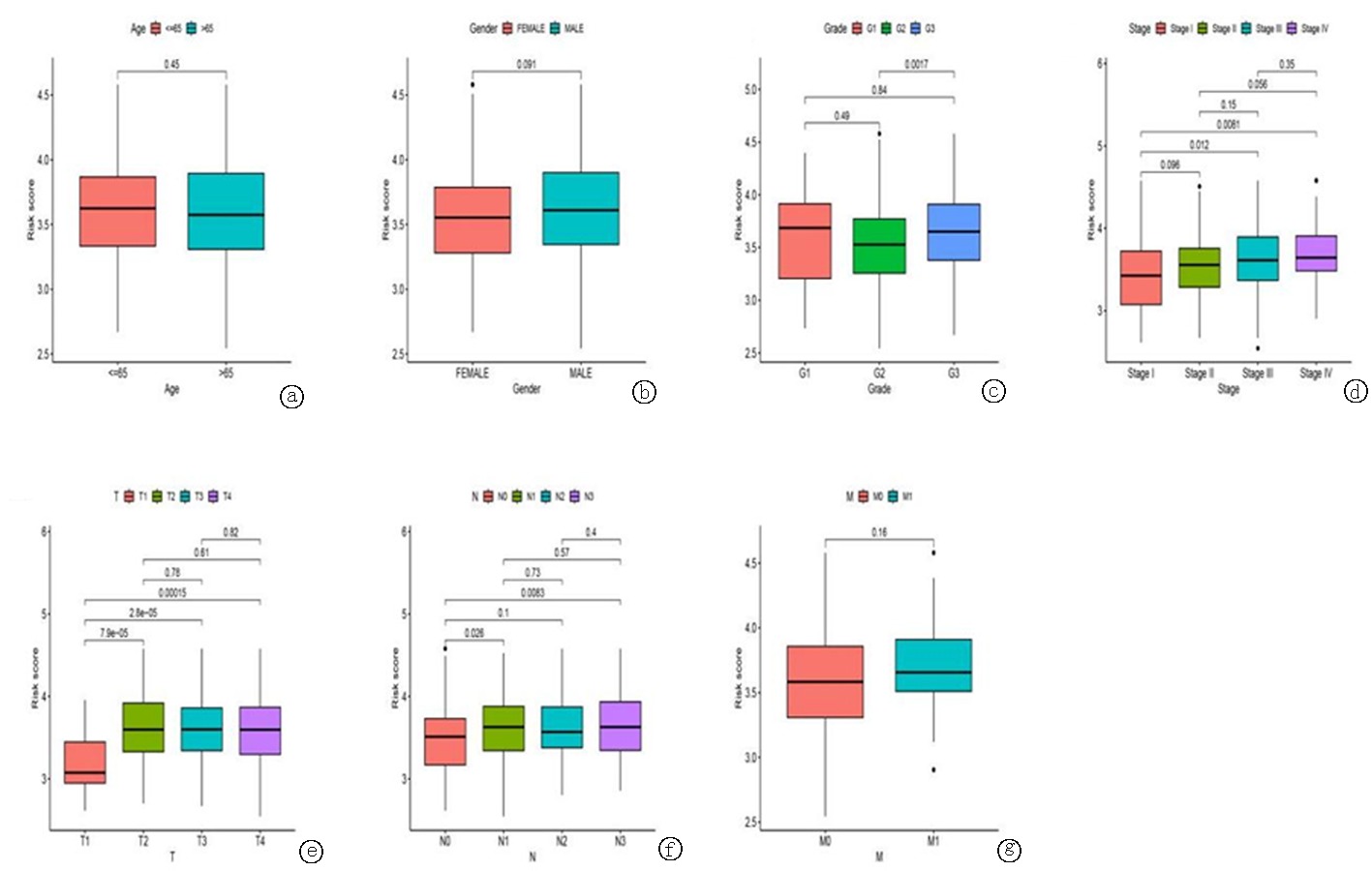
Clinical Focus ›› 2024, Vol. 39 ›› Issue (1): 20-29.doi: 10.3969/j.issn.1004-583X.2024.01.003
Previous Articles Next Articles
Screening of glycolysis-related genes for predicting the prognosis of patients with gastric cancer: Based on bioinformatics
Zhao Xuhui1, Huang Xiaomin1, Da Dezhuan2, Xu Yan1, Cui Xiaodong1, Li Hongling2( )
)
- 1. First School of Clinical Medical, Gansu University of Chinese Medicine, Lanzhou 730000, China
2. Department of Oncology, Gansu Provincial Hospital, Lanzhou 730000, China
-
Received:2023-06-17Online:2024-01-20Published:2024-03-22
CLC Number:
Cite this article
Zhao Xuhui, Huang Xiaomin, Da Dezhuan, Xu Yan, Cui Xiaodong, Li Hongling. Screening of glycolysis-related genes for predicting the prognosis of patients with gastric cancer: Based on bioinformatics[J]. Clinical Focus, 2024, 39(1): 20-29.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://huicui.hebmu.edu.cn/EN/10.3969/j.issn.1004-583X.2024.01.003
| 序号 | 基因 | 离散系数 |
|---|---|---|
| 1 | -0.0962952004774155 | |
| 2 | -0.129632577184322 | |
| 3 | -0.0519404248232357 | |
| 4 | 0.239072070967019 | |
| 5 | 0.147337518442243 | |
| 6 | -0.0836373085400017 | |
| 7 | -0.0370752619271387 | |
| 8 | 0.121593689211912 | |
| 9 | 0.0135857127689336 | |
| 10 | -0.0113107594377815 | |
| 11 | 0.0872242032080347 | |
| 12 | 0.114552783094288 | |
| 13 | 0.891795946885758 | |
| 14 | -0.0217432055166958 | |
| 15 | -0.093175891304856 |
Tab.1 Screened 15 glycolysis-related genes for predicting the prognosis of GC patients using LASSO regression analysis
| 序号 | 基因 | 离散系数 |
|---|---|---|
| 1 | -0.0962952004774155 | |
| 2 | -0.129632577184322 | |
| 3 | -0.0519404248232357 | |
| 4 | 0.239072070967019 | |
| 5 | 0.147337518442243 | |
| 6 | -0.0836373085400017 | |
| 7 | -0.0370752619271387 | |
| 8 | 0.121593689211912 | |
| 9 | 0.0135857127689336 | |
| 10 | -0.0113107594377815 | |
| 11 | 0.0872242032080347 | |
| 12 | 0.114552783094288 | |
| 13 | 0.891795946885758 | |
| 14 | -0.0217432055166958 | |
| 15 | -0.093175891304856 |
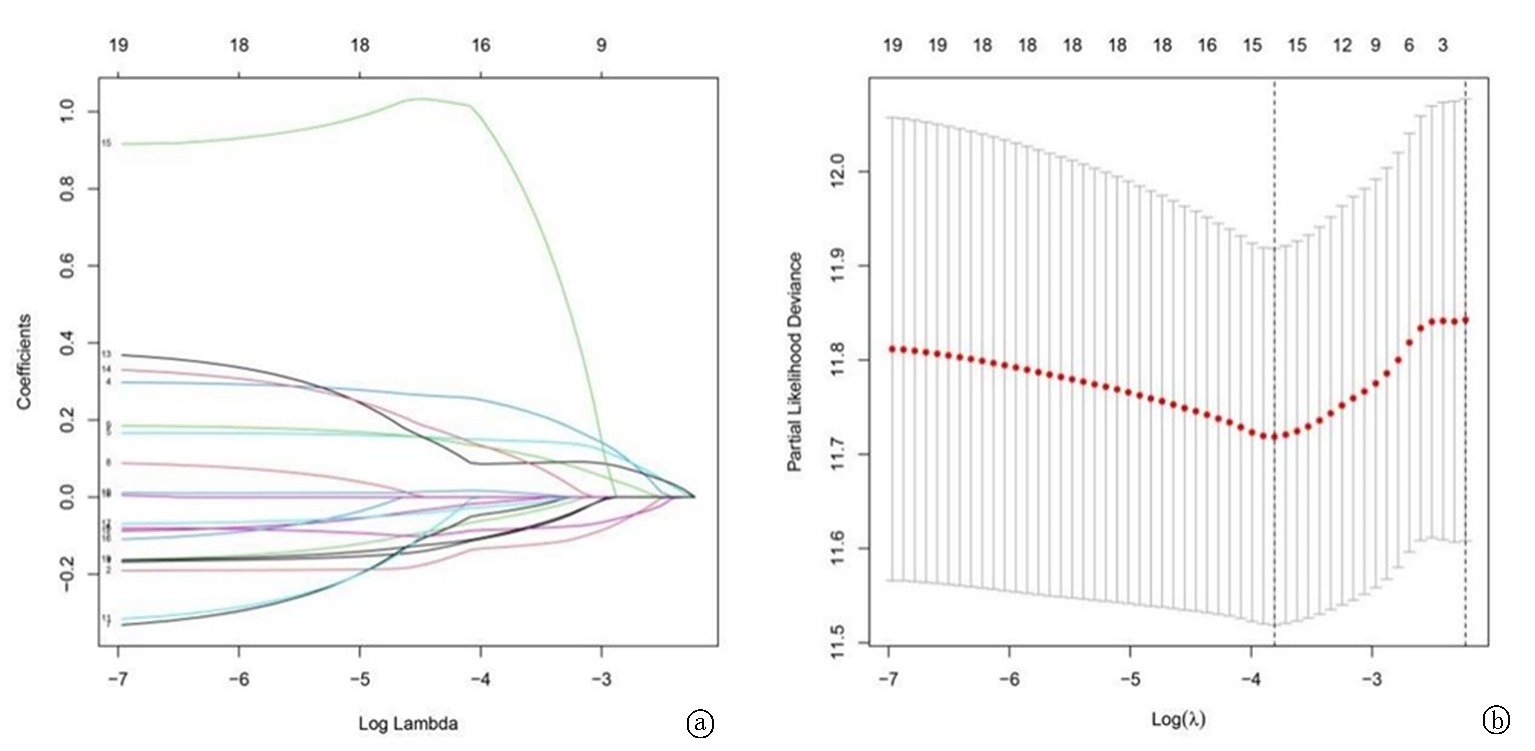
Fig. 2 LASSO regression analysis for construction of the final prediction model a.Determine the optimal parameter(λ) for the LASSO model; plot a vertical dashed line at the optimal value using the minimum criterion; b.Partial likelihood deviance of the LASSO distribution

Fig.5 Nomogram considering glycolysis and clinical factors a.Nomogram illustrating age and risk score for predicting 1-year, 3-year, and 5-year OS; b.Colibration plot of nomogram; c.ROC curves of risk, nomogram and other clinical features
| [1] |
Thrift AP, El-Serag HB. Burden of gastric cancer[J]. Clin Gastroenterol Hepatol, 2020, 18(3):534-542.
doi: 10.1016/j.cgh.2019.07.045 URL |
| [2] |
Sexton RE, Al Hallak MN, Diab M, et al. Gastric cancer: A comprehensive review of current and future treatment strategies[J]. Cancer Metastasis Rev, 2020, 39(4):1179-1203.
doi: 10.1007/s10555-020-09925-3 |
| [3] |
Kang YK, Boku N, Satoh T, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): A randomised, double-blind, placebo-controlled, phase 3 trial[J]. Lancet, 2017, 390(10111):2461-2471.
doi: 10.1016/S0140-6736(17)31827-5 URL |
| [4] | Kang JH, Lee SI, Lim DH, et al. Salvage chemotherapy for pretreated gastric cancer: A randomized phase III trial comparing chemotherapy plus best supportive care with best supportive care alone[J]. Clin Oncol, 2012, 30(13):1513-1518. |
| [5] |
Chen LT, Satoh T, Ryu MH, et al. A phase 3 study of nivolumab in previously treated advanced gastric or gastroesophageal junction cancer (ATTRACTION-2): 2-year update data[J]. Gastric Cancer, 2020, 23(3):510-519.
doi: 10.1007/s10120-019-01034-7 |
| [6] |
Muro K, Chung HC, Shankaran V, et al. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): A multicentre, open-label, phase 1b trial[J]. Lancet Oncol, 2016, 17(6):717-726.
doi: S1470-2045(16)00175-3 pmid: 27157491 |
| [7] |
Vaupel P, Schmidberger H, Mayer A. The Warburg effect: Essential part of metabolic reprogramming and central contributor to cancer progression[J]. Int J Radiat Biol, 2019, 95(7):912-919.
doi: 10.1080/09553002.2019.1589653 pmid: 30822194 |
| [8] |
Dhup S, Dadhich RK, Porporato PE, et al. Multiple biological activities of lactic acid in cancer: Influences on tumor growth, angiogenesis and metastasis[J]. Curr Pharm Des, 2012, 18(10):1319-1330.
doi: 10.2174/138161212799504902 URL |
| [9] |
Hu H, Juvekar A, Lyssiotis CA, et al. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton[J]. Cell, 2016, 164(3):433-446.
doi: 10.1016/j.cell.2015.12.042 pmid: 26824656 |
| [10] | Massari F, Ciccarese C, Santoni M, et al. Metabolic phenotype of bladder cancer[J]. Cancer Treat Rev, 2016,45:46-57. |
| [11] | Lv Z, Qi L, Hu X, et al. Identification of a novel glycolysis-related gene signature correlates with the prognosis and therapeutic responses in patients with clear cell renal cell carcinoma[J]. Front Oncol, 2021,11:633950. |
| [12] |
Liu C, Li Y, Wei M, et al. Identification of a novel glycolysis-related gene signature that can predict the survival of patients with lung adenocarcinoma[J]. Cell Cycle, 2019, 18(5):568-579.
doi: 10.1080/15384101.2019.1578146 pmid: 30727821 |
| [13] | Jiang L, Zhao L, Bi J, et al. Glycolysis gene expression profilings screen for prognostic risk signature of hepatocellular carcinoma[J]. Aging (Albany NY), 2019, 11(23):10861-10882. |
| [14] |
Ruan Y, Tang Q, Qiao J, et al. Identification of a novel glycolysis-related prognosis risk signature in triple-negative breast cancer[J]. Front Oncol, 2023, 13:1171496.
doi: 10.3389/fonc.2023.1171496 URL |
| [15] |
He M, Hu C, Deng J, et al. Identification and validation of a novel glycolysis-related gene signature for predicting the prognosis and therapeutic response in triple-negative breast cancer[J]. Adv Ther, 2023, 40: 310-330.
doi: 10.1007/s12325-022-02330-y |
| [16] |
Yu J, Liu TT, Liang LL, et al. Identification and validation of a novel glycolysis-related gene signature for predicting the prognosis in ovarian cancer[J]. Cancer Cell Int, 2021, 21: 353.
doi: 10.1186/s12935-021-02045-0 pmid: 34229669 |
| [17] |
Zhu J, Wang S, Bai H, et al. Identification of a novel glycolysis-related gene signature for predicting the survival of patients with colon adenocarcinoma[J]. Scand J Gastroenterol, 2022, 57(2):214-221.
doi: 10.1080/00365521.2021.1989026 URL |
| [18] |
Yu S, Hu C, Cai L, et al. Seven-gene signature based on glycolysis is closely related to the prognosis and tumor immune infiltration of patients with gastric cancer[J]. Front Oncol, 2020, 10:1778.
doi: 10.3389/fonc.2020.01778 pmid: 33072557 |
| [19] |
Liu Y, Wu M, Cao J, et al. Identification and verification of a glycolysis-related gene signature for gastric cancer[J]. Ann Transl Med, 2022, 10: 1010.
doi: 10.21037/atm-22-3980 pmid: 36267782 |
| [20] | 陈艳昕, 刘庆滨, 黄江梅, 等. 葡萄糖转运蛋白1在胃癌的表达及临床病理意义[J]. 临床荟萃, 2012, 27(8):694-695. |
| [21] |
Nehring H, Meierjohann S, Friedmann Angeli JP. Emerging aspects in the regulation of ferroptosis[J]. Biochem Soc Trans, 2020, 48(5):2253-2259.
doi: 10.1042/BST20200523 URL |
| [22] |
Vaupel P, Multhoff G. Revisiting the Warburg effect: Historical dogma versus current understanding[J]. J Physiol, 2021, 599(6):1745-1757.
doi: 10.1113/tjp.v599.6 URL |
| [23] | Hu Q, Qin Y, Ji S, et al. UHRF1 promotes aerobic glycolysis and proliferation via suppression of SIRT4 in pancreatic cancer[J]. Cancer Lett, 2019,452:226-236. |
| [24] |
Zhao S, Guan B, Mi Y, et al. LncRNA MIR17HG promotes colorectal cancer liver metastasis by mediating a glycolysis-associated positive feedback circuit[J]. Oncogene, 2021, 40(28):4709-4724.
doi: 10.1038/s41388-021-01859-6 pmid: 34145399 |
| [25] |
Cai L, Ye Y, Jiang Q, et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma[J]. Nat Commun, 2015, 6: 7353.
doi: 10.1038/ncomms8353 pmid: 26135619 |
| [26] |
Iizasa H, Kim H, Kartika AV, et al. Corrigendum: Role of viral and host microRNAs in immune regulation of Epstein-Barr virus-associated diseases[J]. Front Immunol, 2020, 11: 498.
doi: 10.3389/fimmu.2020.00498 pmid: 32318060 |
| [27] | 程燕妮, 刘佳, 袁野, 等. 丙酮酸激酶M2型在恶性肿瘤中的表达及其临床检测研究进展[J]. 临床荟萃, 2022, 37(3):279-284. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||