Clinical Focus ›› 2022, Vol. 37 ›› Issue (11): 977-984.doi: 10.3969/j.issn.1004-583X.2022.11.003
Previous Articles Next Articles
MDM4rs4245739 gene polymorphism and breast cancer susceptibility:A meta-analysis
Jiang Yan1a,b, Li Jiayang1c, Wu Hongyu2, Chen Baolin1a,b, Cheng Xiaoming1a,b, Lyu Junyuan1a,b( )
)
- 1. Department of General Surgery; b.Department of Breast and Thyroid Surgery; c.Drug Clinical Trial Institution,the Affiliated Hospital of Zunyi Medical University,Zunyi 563099,China
2. Key Laboratory of Basic Pharmacology of Ministry of Education and Joint International Research Laboratory of Ethnomedicine of Ministry of Education,Zunyi Medical University,Zunyi 563000,China
-
Received:2022-03-14Online:2022-11-20Published:2023-01-02 -
Contact:Lyu Junyuan E-mail:junyuanlv@zmu.edu.cn
CLC Number:
Cite this article
Jiang Yan, Li Jiayang, Wu Hongyu, Chen Baolin, Cheng Xiaoming, Lyu Junyuan. MDM4rs4245739 gene polymorphism and breast cancer susceptibility:A meta-analysis[J]. Clinical Focus, 2022, 37(11): 977-984.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://huicui.hebmu.edu.cn/EN/10.3969/j.issn.1004-583X.2022.11.003
| 第一作者 | 年份 | 地区 | 种族 | 病例/对照数 | 病例组 | 对照组 | SOC | HWE | NOS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | AA | AC | CC | |||||||||
| Liu et al[ | 2013 | 中国 | 非高加索人 | 800/800 | 733 | 67 | 0 | 686 | 111 | 3 | PB | 0.505 | 6 | |
| Liu et al[ | 2013 | 中国 | 非高加索人 | 300/600 | 278 | 22 | 0 | 501 | 96 | 3 | PB | 0.483 | 6 | |
| Hashemi et al[ | 2018 | 伊朗 | 非高加索人 | 265/221 | 175 | 83 | 7 | 142 | 70 | 9 | HB | 0.919 | 5 | |
| Montserrat et al[ | 2013 | 混合 | 高加索人 | 6 512/41 451 | 3 318 | 2 637 | 557 | 22 825 | 15 798 | 2 828 | Mixed | 0.183 | 5 | |
| Gansmo et al[ | 2015 | 挪威 | 高加索人 | 1 717/1 870 | 966 | 643 | 108 | 1 021 | 703 | 146 | PB | 0.106 | 6 | |
| Pedram et al[ | 2016 | 伊朗 | 非高加索人 | 220/260 | 123 | 87 | 10 | 165 | 81 | 14 | HB | 0.335 | 6 | |
| 第一作者 | 年份 | 地区 | 种族 | 病例/对照数 | 病例组 | 对照组 | SOC | HWE | NOS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | AA | AC | CC | |||||||||
| Liu et al[ | 2013 | 中国 | 非高加索人 | 800/800 | 733 | 67 | 0 | 686 | 111 | 3 | PB | 0.505 | 6 | |
| Liu et al[ | 2013 | 中国 | 非高加索人 | 300/600 | 278 | 22 | 0 | 501 | 96 | 3 | PB | 0.483 | 6 | |
| Hashemi et al[ | 2018 | 伊朗 | 非高加索人 | 265/221 | 175 | 83 | 7 | 142 | 70 | 9 | HB | 0.919 | 5 | |
| Montserrat et al[ | 2013 | 混合 | 高加索人 | 6 512/41 451 | 3 318 | 2 637 | 557 | 22 825 | 15 798 | 2 828 | Mixed | 0.183 | 5 | |
| Gansmo et al[ | 2015 | 挪威 | 高加索人 | 1 717/1 870 | 966 | 643 | 108 | 1 021 | 703 | 146 | PB | 0.106 | 6 | |
| Pedram et al[ | 2016 | 伊朗 | 非高加索人 | 220/260 | 123 | 87 | 10 | 165 | 81 | 14 | HB | 0.335 | 6 | |
| 分组 | 数量 | C vs A | AC+CC vs AA | AC+AA vs CC | AC vs AA | CC vs AA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 种族 | |||||||||||||||
| 非高加索人 | 4 | 0.70(0.46-1.06)* | 0.093 | 0.74(0.44-1.22)* | 0.236 | 0.67(0.36-1.24)# | 0.200 | 0.76(0.45-1.28)* | 0.302 | 0.71(0.38-1.32)# | 0.279 | ||||
| 高加索人 | 2 | 1.04(0.83-1.30)* | 0.753 | 1.06(0.84-1.33)* | 0.639 | 1.02(0.64-1.63)* | 0.923 | 1.06(0.90-1.26)* | 0.478 | 1.04(0.61-1.79)* | 0.876 | ||||
| 地区 | |||||||||||||||
| 亚洲 | 4 | 0.70(0.46-1.06)* | 0.093 | 0.74(0.44-1.22)* | 0.236 | 0.67(0.36-1.24)# | 0.200 | 0.76(0.45-1.28)* | 0.302 | 0.71(0.38-1.32)# | 0.279 | ||||
| 欧洲 | 1 | 0.92(0.83-1.02) | 0.116 | 0.93(0.82-1.06) | 0.292 | 0.79(0.61-1.03) | 0.080 | 0.96(0.84-1.11) | 0.590 | 0.78(0.60-1.02) | 0.068 | ||||
| 混合 | 1 | 1.16(1.11-1.21) | 0.000 | 1.18(1.12-1.24) | 0.000 | 1.28(1.16-1.41) | 0.000 | 1.15(1.09-1.21) | 0.000 | 1.36(1.23-1.49) | 0.000 | ||||
| 样本量 | |||||||||||||||
| >1 000 | 2 | 1.04(0.83-1.30)* | 0.753 | 1.06(0.84-1.33)* | 0.639 | 1.02(0.64-1.63)* | 0.923 | 1.06(0.90-1.26)* | 0.478 | 1.04(0.61-1.79)* | 0.876 | ||||
| <1 000 | 4 | 0.70(0.46-1.06)* | 0.093 | 0.74(0.44-1.22)* | 0.236 | 0.67(0.36-1.24)# | 0.200 | 0.76(0.45-1.28)* | 0.302 | 0.71(0.38-1.32)# | 0.279 | ||||
| 基因分型检测方法 | |||||||||||||||
| PCR-RFLP | 4 | 0.70(0.46-1.06)* | 0.093 | 0.74(0.44-1.22)* | 0.236 | 0.67(0.36-1.24)# | 0.200 | 0.76(0.45-1.28)* | 0.302 | 0.71(0.38-1.32)# | 0.279 | ||||
| 其他 | 2 | 1.04(0.83-1.30)* | 0.753 | 1.06(0.84-1.33)* | 0.639 | 1.02(0.64-1.24)* | 0.923 | 1.06(0.90-1.26)* | 0.478 | 1.04(0.61-1.79)* | 0.876 | ||||
| 健康对照者来源 | |||||||||||||||
| PB | 3 | 0.62(0.38-1.01)* | 0.054 | 0.61(0.37-1.02)* | 0.060 | 0.78(0.60-1.01)# | 0.054 | 0.63(0.38-1.05)* | 0.079 | 0.77(0.59-0.99)# | 0.044 | ||||
| HB | 2 | 1.00(0.80-1.25)# | 0.976 | 1.13(0.77-1.66)* | 0.542 | 0.75(0.40-1.42)# | 0.378 | 1.18(0.79-1.75)* | 0.415 | 0.81(0.42-1.54)# | 0.518 | ||||
| 混合 | 1 | 1.16(1.11-1.21) | 0.000 | 1.18(1.12-1.24) | 0.000 | 1.28(1.16-1.41) | 0.000 | 1.15(1.09-1.21) | 0.000 | 1.35(1.23-1.49) | 0.000 | ||||
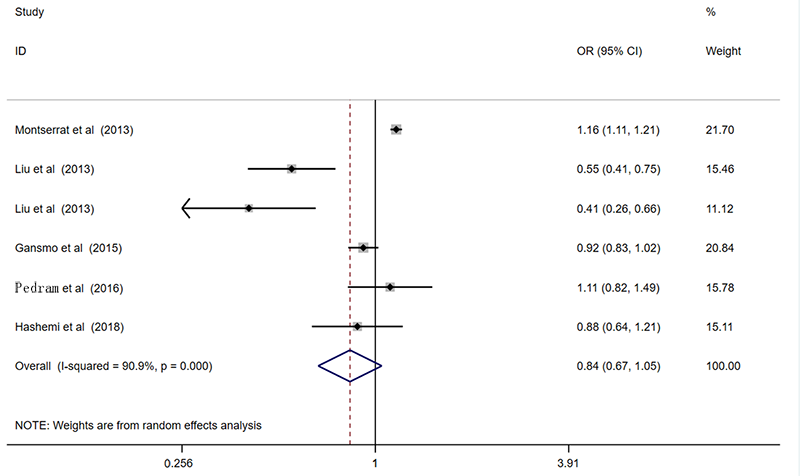
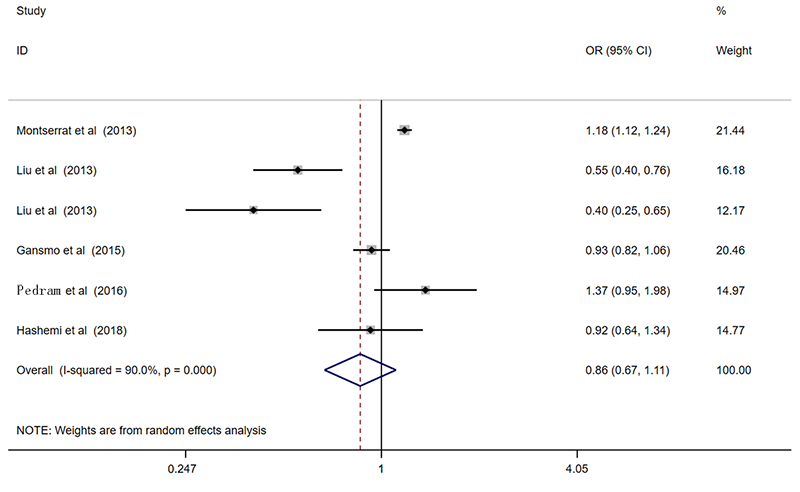
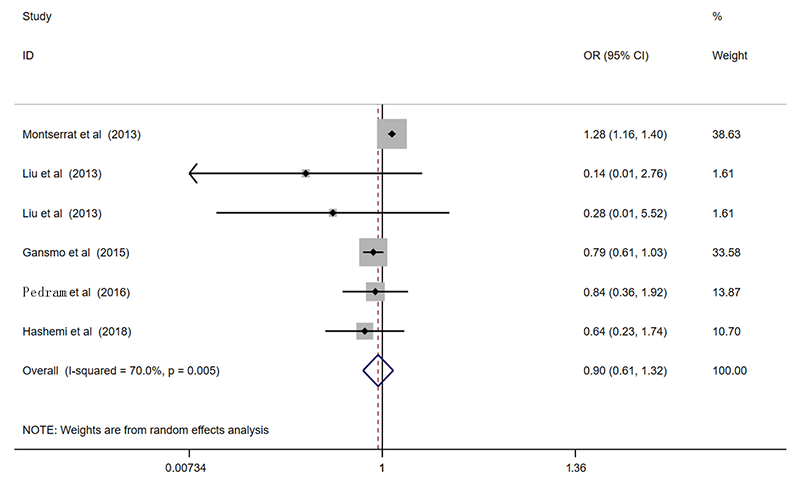
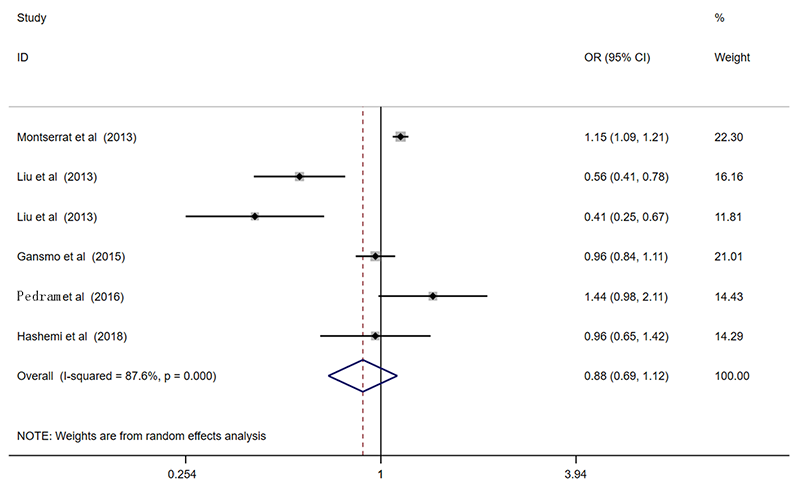
| 总共 | 6 | 0.84(0.67-1.05)* | 0.118 | 0.86(0.67-1.11)* | 0.245 | 0.90(0.61-1.32)* | 0.585 | 0.88(0.69-1.12)* | 0.305 | 0.90(0.59-1.40)* | 0.649 | ||||
| 分组 | 数量 | C vs A | AC+CC vs AA | AC+AA vs CC | AC vs AA | CC vs AA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 种族 | |||||||||||||||
| 非高加索人 | 4 | 0.70(0.46-1.06)* | 0.093 | 0.74(0.44-1.22)* | 0.236 | 0.67(0.36-1.24)# | 0.200 | 0.76(0.45-1.28)* | 0.302 | 0.71(0.38-1.32)# | 0.279 | ||||
| 高加索人 | 2 | 1.04(0.83-1.30)* | 0.753 | 1.06(0.84-1.33)* | 0.639 | 1.02(0.64-1.63)* | 0.923 | 1.06(0.90-1.26)* | 0.478 | 1.04(0.61-1.79)* | 0.876 | ||||
| 地区 | |||||||||||||||
| 亚洲 | 4 | 0.70(0.46-1.06)* | 0.093 | 0.74(0.44-1.22)* | 0.236 | 0.67(0.36-1.24)# | 0.200 | 0.76(0.45-1.28)* | 0.302 | 0.71(0.38-1.32)# | 0.279 | ||||
| 欧洲 | 1 | 0.92(0.83-1.02) | 0.116 | 0.93(0.82-1.06) | 0.292 | 0.79(0.61-1.03) | 0.080 | 0.96(0.84-1.11) | 0.590 | 0.78(0.60-1.02) | 0.068 | ||||
| 混合 | 1 | 1.16(1.11-1.21) | 0.000 | 1.18(1.12-1.24) | 0.000 | 1.28(1.16-1.41) | 0.000 | 1.15(1.09-1.21) | 0.000 | 1.36(1.23-1.49) | 0.000 | ||||
| 样本量 | |||||||||||||||
| >1 000 | 2 | 1.04(0.83-1.30)* | 0.753 | 1.06(0.84-1.33)* | 0.639 | 1.02(0.64-1.63)* | 0.923 | 1.06(0.90-1.26)* | 0.478 | 1.04(0.61-1.79)* | 0.876 | ||||
| <1 000 | 4 | 0.70(0.46-1.06)* | 0.093 | 0.74(0.44-1.22)* | 0.236 | 0.67(0.36-1.24)# | 0.200 | 0.76(0.45-1.28)* | 0.302 | 0.71(0.38-1.32)# | 0.279 | ||||
| 基因分型检测方法 | |||||||||||||||
| PCR-RFLP | 4 | 0.70(0.46-1.06)* | 0.093 | 0.74(0.44-1.22)* | 0.236 | 0.67(0.36-1.24)# | 0.200 | 0.76(0.45-1.28)* | 0.302 | 0.71(0.38-1.32)# | 0.279 | ||||
| 其他 | 2 | 1.04(0.83-1.30)* | 0.753 | 1.06(0.84-1.33)* | 0.639 | 1.02(0.64-1.24)* | 0.923 | 1.06(0.90-1.26)* | 0.478 | 1.04(0.61-1.79)* | 0.876 | ||||
| 健康对照者来源 | |||||||||||||||
| PB | 3 | 0.62(0.38-1.01)* | 0.054 | 0.61(0.37-1.02)* | 0.060 | 0.78(0.60-1.01)# | 0.054 | 0.63(0.38-1.05)* | 0.079 | 0.77(0.59-0.99)# | 0.044 | ||||
| HB | 2 | 1.00(0.80-1.25)# | 0.976 | 1.13(0.77-1.66)* | 0.542 | 0.75(0.40-1.42)# | 0.378 | 1.18(0.79-1.75)* | 0.415 | 0.81(0.42-1.54)# | 0.518 | ||||
| 混合 | 1 | 1.16(1.11-1.21) | 0.000 | 1.18(1.12-1.24) | 0.000 | 1.28(1.16-1.41) | 0.000 | 1.15(1.09-1.21) | 0.000 | 1.35(1.23-1.49) | 0.000 | ||||
| 总共 | 6 | 0.84(0.67-1.05)* | 0.118 | 0.86(0.67-1.11)* | 0.245 | 0.90(0.61-1.32)* | 0.585 | 0.88(0.69-1.12)* | 0.305 | 0.90(0.59-1.40)* | 0.649 | ||||
| 协变量 | N | C vs A | AC+CC vs AA | AC+AA vs CC | AC vs AA | CC vs AA |
|---|---|---|---|---|---|---|
| 种族 | 2 | 0.201 | 0.341 | 0.244 | 0.388 | 0.301 |
| 地区 | 3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 样本量 | 2 | 0.201 | 0.341 | 0.244 | 0.388 | 0.301 |
| 基因分型检测方法 | 2 | 0.201 | 0.341 | 0.244 | 0.388 | 0.301 |
| 健康对照者来源 | 3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 协变量 | N | C vs A | AC+CC vs AA | AC+AA vs CC | AC vs AA | CC vs AA |
|---|---|---|---|---|---|---|
| 种族 | 2 | 0.201 | 0.341 | 0.244 | 0.388 | 0.301 |
| 地区 | 3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 样本量 | 2 | 0.201 | 0.341 | 0.244 | 0.388 | 0.301 |
| 基因分型检测方法 | 2 | 0.201 | 0.341 | 0.244 | 0.388 | 0.301 |
| 健康对照者来源 | 3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| [1] |
Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 71(3): 209-249.
doi: 10.3322/caac.21660 URL |
| [2] |
Lilyquist J, Ruddy KJ, Vachon CM, et al. Common genetic variation and breast cancer risk-past, present, and future[J]. Cancer Epidemiol Biomarkers Prev, 2018, 27(4): 380-394.
doi: 10.1158/1055-9965.EPI-17-1144 URL |
| [3] |
Rudolph A, Chang-Claude J, Schmidt MK. Gene-environment interaction and risk of breast cancer[J]. Br J Cancer, 2016, 114(4): 125-133.
doi: 10.1038/bjc.2015.439 URL |
| [4] |
Rahman N. Realizing the promise of cancer predisposition genes[J]. Nature, 2014, 505(7483): 302-308.
doi: 10.1038/nature12981 URL |
| [5] |
Haupt S, Vijayakumaran R, Miranda PJ, et al. The role of MDM2 and MDM4 in breast cancer development and prevention[J]. J Mol Cell Biol, 2017, 9(1):53-61.
doi: 10.1093/jmcb/mjx007 pmid: 28096293 |
| [6] |
Stegeman S, Moya L, Selth LA, et al. A genetic variant of MDM4 influences regulation by multiple microRNAs in prostate cancer[J]. Endocr Relat Cancer, 2015, 22(2): 265-276.
doi: 10.1530/ERC-15-0013 URL |
| [7] |
Liu J, Tang X, Li M, et al. Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility[J]. Breast Cancer Res Treat, 2013, 140(1): 151-157.
doi: 10.1007/s10549-013-2615-x URL |
| [8] |
Hashemi M, Sanaei S, Hashemi SM, et al. Association of single nucleotide polymorphisms of the MDM4 gene with the susceptibility to breast cancer in a Southeast Iranian population sample[J]. Clin Breast Cancer, 2018, 18(5): e883-e891.
doi: 10.1016/j.clbc.2018.01.003 URL |
| [9] | Wang YX, Yang Z, Chang XL, et al. Five MDM4 gene polymorphisms on cancer risk: An updated systematic review and meta-analysis[J]. Int J Biol Markers, 2021, 36(2): 17246008211033874. |
| [10] |
Montserrat GC, Couch FJ, Lindstrom S, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci[J]. Nat Genet, 2013, 45(4): 392-398e2.
doi: 10.1038/ng.2561 URL |
| [11] |
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses[J]. Eur J Epidemiol, 2010, 25(9): 603-605.
doi: 10.1007/s10654-010-9491-z pmid: 20652370 |
| [12] |
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis[J]. Stat Med, 2002, 21(11): 1539-1558.
doi: 10.1002/sim.1186 pmid: 12111919 |
| [13] |
Peters JL, Sutton AJ, Jones DR, et al. Comparison of two methods to detect publication bias in meta-analysis[J]. JAMA, 2006, 295(6): 676-680.
doi: 10.1001/jama.295.6.676 pmid: 16467236 |
| [14] | Jin X, Zhao W, Zheng M, et al. The role of MDM4 SNP34091 A>C polymorphism in cancer: A meta-analysis on 19, 328 patients and 51, 058 controls[J]. Int J Biol Markers, 2017, 32(1): e62-e67. |
| [15] |
Wang MJ, Luo Y J, Shi ZY, et al. The associations between MDM4 gene polymorphisms and cancer risk[J]. Oncotarget, 2016, 7(34): 55611-55623.
doi: 10.18632/oncotarget.10877 URL |
| [16] |
Zhai Y, Dai Z, He H, et al. A PRISMA-compliant meta-analysis of MDM4 genetic variants and cancer susceptibility[J]. Oncotarget, 2016, 7(45): 73935-73944.
doi: 10.18632/oncotarget.12558 pmid: 27738340 |
| [17] | Song CG, Fu FM, Wu XY, et al. Correlation of polymorphism rsl563828 in MDM4 gene with breast cancer risk and onset age[J]. Chin J Surg, 2012, 50(1): 53-56. |
| [18] |
Atwal GS, Kirchhoff T, Bond EE, et al. Altered tumor formation and evolutionary selection of genetic variants in the human MDM4 oncogene[J]. Proc Natl Acad Sci USA, 2009, 106(25): 10236-10241.
doi: 10.1073/pnas.0901298106 URL |
| [19] |
Bauer M, Kantelhardt EJ, Stiewe T, et al. Specific allelic variants of SNPs in the MDM2 and MDMX genes are associated with earlier tumor onset and progression in Caucasian breast cancer patients[J]. Oncotarget, 2019, 10(20): 1975-1992.
doi: 10.18632/oncotarget.26768 URL |
| [20] |
Vulsteke C, Lambrechts D, Dieudonne A, et al. Genetic variability in the multidrug resistance associated protein-1 (ABCC1/MRP1) predicts hematological toxicity in breast cancer patients receiving (neo-)adjuvant chemotherapy with 5-fluorouracil, epirubicin and cyclophosphamide (FEC)[J]. Ann Oncol, 2013, 24(6): 1513-1525.
doi: 10.1093/annonc/mdt008 pmid: 23396606 |
| [21] |
Kulkarni DA, Vazquez A, Haffty BG, et al. A polymorphic variant in human MDM4 associates with accelerated age of onset of estrogen receptor negative breast cancer[J]. Carcinogenesis, 2009, 30(11): 1910-1915.
doi: 10.1093/carcin/bgp224 pmid: 19762336 |
| [22] | Joseph V, Gaudet M, Devlin V, et al. Genetic variation of human MDM4 confers increased risk for breast cancer in the Ashkenazi Jewish population[J]. Curr Oncol, 2009, 16(5): 97. |
| [23] |
Reincke S, Govbakh L, Wilhelm B, et al. Mutation analysis of the MDM4 gene in German breast cancer patients[J]. BMC cancer, 2008, 8: 52.
doi: 10.1186/1471-2407-8-52 pmid: 18279506 |
| [24] |
Bartnykaite A, Savukaityte A, Ugenskiene R, et al. Associations of MDM2 and MDM4 polymorphisms with early-stage breast cancer[J]. J Clin Med, 2021, 10(4): 866.
doi: 10.3390/jcm10040866 URL |
| [25] |
Gansmo LB, Romundstad P, Birkeland E, et al. MDM4 SNP34091 (rs4245739) and its effect on breast-, colon-, lung-, and prostate cancer risk[J]. Cancer med, 2015, 4(12): 1901-1907.
doi: 10.1002/cam4.555 URL |
| [26] |
Pedram N, Pouladi N, Feizi MA, et al. Analysis of the Association between MDM4 rs4245739 single nucleotide polymorphism and breast cancer susceptibility[J]. Clin lab, 2016, 62(7): 1303-1308.
doi: 10.7754/Clin.Lab.2016.151128 pmid: 28164646 |
| [27] |
Dai X, Li T, Bai Z, et al. Breast cancer intrinsic subtype classification, clinical use and future trends[J]. Am J Cancer Res, 2015, 5(10): 2929-2943.
pmid: 26693050 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||